Article Text
Abstract
Background Pseudomonas aeruginosa frequently colonises intubated patients and causes life-threatening ventilator-associated pneumonia (VAP). The role of quorum sensing (QS), regulating virulence in this pathogen, during colonisation and development of VAP is unknown.
Methods P aeruginosa isolates and tracheal aspirates were prospectively collected from intubated patients. Genotypes and QS-independent virulence traits (exoU, exoS, PAPI-1 and PAPI-2) harboured by colonising isolates were identified in vitro with the CLONDIAG array. The production of elastase and rhamnolipids was measured to assess QS-dependent virulence. To monitor QS activity ‘in patient’, total RNA was extracted directly from tracheal aspirates and expression of QS genes was measured.
Results 320 P aeruginosa isolates and tracheal aspirates were obtained from 29 patients of whom 6 developed VAP (20%). Seven patients (24%) were initially colonised by QS-proficient isolates; 57% of them developed VAP as compared with 9% of patients colonised by QS-deficient isolates (p=0.018). Of all tested virulence traits from the initial colonising isolates, only rhamnolipids were associated with development of VAP (p=0.003). VAP occurred more frequently in patients colonised during the entire observation period by isolates producing high levels of rhamnolipids (p=0.001). ‘In patient’ monitoring of QS genes showed non-induced expression profiles in patients without VAP. In contrast, exponential induction of QS circuit and target gene expression was observed for two patients with VAP, and an ‘in patient’ QS gene expression profile and hierarchy similar to those in vitro was measured for one patient with VAP.
Conclusions Production of the QS-dependent virulence factor rhamnolipids by colonising P aeruginosa isolates is associated with development of VAP.
- Pseudomonas aeruginosa
- ventilator-associated pneumonia
- colonisation
- quorum sensing
- virulence
- bacterial infection
- pneumonia
- respiratory infection
Statistics from Altmetric.com
- Pseudomonas aeruginosa
- ventilator-associated pneumonia
- colonisation
- quorum sensing
- virulence
- bacterial infection
- pneumonia
- respiratory infection
Introduction
Pseudomonas aeruginosa frequently colonises intubated patients. In 10–20% of these patients colonisation progresses to ventilator-associated pneumonia (VAP) associated with mortality rates of 30–40%.1 To date, it has been impossible to accurately identify those colonised patients who will develop this complication. In particular it remains unclear whether patients prone to infection are colonised by especially virulent strains. A quorum-sensing (QS) circuit regulates most virulence determinants in P aeruginosa according to cell density.2–6 These include many secreted factors (elastase, phospholipase C, lecitinase, rhamnolipids) as well as secondary metabolites (pyocyanin, cyanide).7 8 The importance of QS-regulated virulence traits for the pathogenicity of P aeruginosa has been established in both animal and plant infection models.9 However, evidence for QS activity in the human host remains scarce and is derived from the detection of QS signalling molecules and the detection of QS gene expression in sputum samples collected occasionally from patients with cystic fibrosis.10–15 Its relevance in acute human infections has even been questioned recently.16 17 If QS-dependent virulence traits were important for the development of infection, then preventive measures aiming at the inhibition of the QS circuit would be an attractive alternative to conventional antibiotics, which frequently lead to the selection of resistance without being able to eradicate colonisation of intubated patients.1 4 18
Our primary aim was to determine the presence of particular virulence traits in P aeruginosa isolates colonising intubated patients and their potential association with the development of P aeruginosa VAP. We also sought to determine whether induction of the QS circuit could be demonstrated ‘in patient’ during colonisation and/or infection of intubated patients by monitoring bacterial gene expression directly in tracheal aspirates.
Methods
Subjects
This study is a subanalysis of a multicentre placebo-controlled trial investigating azithromycin as a QS inhibitor for the prevention of P aeruginosa VAP in colonised mechanically ventilated patients (http://ClinicalTrials.gov ID# NCT00610623) . Detailed information on the results of this clinical trial are published elsewhere (van Delden et al, submitted, 2010). Patients of the present study were restrictively selected from the placebo-control group on the basis of availability of both daily clinical P aeruginosa isolates and tracheal aspirates. We screened intubated patients for respiratory tract colonisation by P aeruginosa every 48 h. Patients with ongoing P aeruginosa infection, or having received antimicrobial treatments active against the colonising isolates during the last 14 days, or during the observation period, were not included. The diagnosis of P aeruginosa VAP was based on the clinical picture, including a pulmonary infection score (CPIS) ≥6, as well as a quantitative culture of a bronchoalveolar lavage fluid yielding >104 CFU/ml of P aeruginosa.19
Collection of respiratory samples
Starting the first day of proven colonisation (D0), we prospectively collected daily tracheal aspirates and one P aeruginosa isolate (collection period: 3–20 days). Samples were frozen at −80°C on site within 15 min, and sent on dry ice to the research laboratory at the University Hospital Geneva, where all RNA and DNA extractions were performed. Tracheal aspirates were used to determine P aeruginosa bacterial loads and ‘in patient’expression of QS genes, whereas P aeruginosa isolates were used for genotypic and phenotypic characterisations (figure 1A). Reasons to stop sample collection were either extubation or proven P aeruginosa VAP.
(A) Flowchart for procedures on tracheal aspirates. From each daily aspirate one Pseudomonas aeruginosa isolate was selected, and DNA as well as RNA were extracted in parallel. Bacterial load was deduced from genomic DNA by quantitative reverse transcription–PCR (qRT–PCR) and gene expression was determined after reverse transcription of total RNA. (B) Bacterial load over time. Shown are the mean±SEM of bacterial loads for patients with ventilator-associated pneumonia (VAP) (filled squares) (n=5, genomic samples of patient 13128 with VAP were not available due to technical problems) and patients without VAP (open squares) (n=23). Arrows indicate days of VAP (dotted arrow indicates patient 13128, for whom no genomic DNA samples were available).
Genomic analysis of clinical isolates
Intrapatient comparison of genotypes was performed on the entire collection of 320 isolates by random amplification of polymorphic DNA (RAPD) using primer 207.20 Interpatient comparison of the genotype of the initial isolate from each of the 29 patients was performed with the P aeruginosa CLONDIAG array.21 The array further detects the presence of 38 genetic markers of the accessory genome, including two genes from the pathogenicity island PAPI-1 and three from PAPI-2.22
The lasR and rhlR genes were amplified by PCR using primers lasR-F1 (5′-ATCTTGTGGGCTGACTGGAC-3′) and lasR8 (5′-CTGCGGCAGTCGTTTCGAGAAT-3′), and rhlR-F1 (5′-ACGGTGCTGGCATAACAGATAGG-3′); and rhlR-R1 (5′-CCTCTCAGTCGGAGGACATACCA-3′), respectively, from bacterial lysates. PCR conditions were as follows: 95°C for 2 min, 27 cycles at 95°C for 15 s, 57°C for 30 s and 72°C for 1 min, and a final extension at 72°C for 4 min. lasR and rhlR PCR products were purified (Qiagen, Hilden, Germany) and sequenced using primers lasR-F1 and rhlR-F1, respectively.
Exoproduct analysis
Elastase activity was determined using the Elastin Congo Red assay in supernatants of cultures grown for 7 h at 37°C in PB medium.23 24 Rhamnolipid production was assessed on modified SW-Blue plates.25 The diameter of the rhamnolipid-containing halo formed around the bacterial colony after 24 h incubation at 37°C and after 24 h at room temperature was measured and compared with that produced by the reference strain PAO1. All determinations were done in duplicate and expressed as a percentage of PAO1 production levels.
DNA and RNA preparation
Genomic DNA, and RNA were extracted from 0.05 ml and 0.5 ml of homogenised tracheal aspirates, respectively (figure 1A). Additional details on the extraction procedures and gene expression analyses are provided in the online data supplement.
Statistical analysis
This study is a subanalysis of a multicentre placebo-controlled trial. None of the analysis presented here was included in the primary or secondary outcomes of this trial, which was focused on the occurrence of P aeruginosa VAP (primary outcome) and mortality (secondary outcome) in both placebo and azithromycin study arms. Two-sided Fisher exact tests were used to analyse differences between group proportions. The Mann–Whitney test was used as a non-parametric test.
Results
Genotypes and virulence phenotypes of initial colonising P aeruginosa isolates
The collection included a total of 320 P aeruginosa isolates from 29 patients. Six patients developed P aeruginosa VAP. Detailed clinical information on the study population is presented in the online data supplement. Four patients (13122, 15101, 21107 and 26102) were colonised by two different genotypes. We first focused on the 29 initial colonising isolates. Using the P aeruginosa CLONDIAG genome array, we distinguished 18 clones (figure 2).21 The CLONDIAG array also allows determination of the presence of virulence genes including the Pseudomonas pathogenicity islands PAPI-1 and PAPI-2, as well as the two mutually exclusive cytotoxicity genes exoS and exoU. The genes of PAPI-1 were detected in only two isolates. In contrast, genes of PAPI-2 were found in 25 out of the 29 isolates (figure 2). The gene encoding the cytotoxic phospholipase ExoU, carried on PAPI-2, was present in isolates of 11 patients (38%), while exoS, not located on a pathogenicity island, was identified in isolates from the other 18 patients (62%). Twenty-two of the initial 29 isolates (76%) produced reduced amounts of the QS-dependent virulence factors elastase and/or rhamnolipids compared with the laboratory strain PAO1 (figure 2). Sequencing revealed that lack of production of these QS-dependent virulence factors correlated for most isolates with mutations in the lasR and/or the rhlR QS-regulator genes (figure 2).26
Phenotypic and genotypic characterization of initial colonising Pseudomonas aeruginosa isolates. Production of elastase (Ela) and rhamnolipids (Rha) was scored according to the classification scheme. Alterations to the PAO1 (wild-type) sequences of the lasR and rhlR genes, as well as undefined deletions (Δ) or insertions (IS) are indicated. The presence of the cytotoxicity genes exoS and exoU from PAPI-1 (maximum of two genes) and PAPI-2 (maximum of three genes), as well as the single nucleotide polymorphism (SNP) type, were established using the CLONDIAG array. Patients with ventilator-associated pneumonia (VAP) are highlighted by black boxes.
We further compared genotypes and virulence phenotypes of the initial colonising isolates from the six patients with VAP with those from the 23 patients without VAP. We found no statistically significant association between VAP and a particular genotype, or the presence of either of the Pseudomonas pathogenicity islands PAPI-1 and PAPI-2, or of the cytotoxicity genes exoU or exoS (table 1). In contrast four out of seven patients (57%) initially colonised by QS-proficient isolates (producing both elastase and rhamnolipids at levels comparable with PAO1) developed VAP as compared with two out of 22 patients (9%) colonised by isolates with reduced QS activities (p=0.018). High level production of rhamnolipids (p=0.003) but not of elastase was statistically associated with VAP (figure 2 and table 1).
Virulence determinants in patients with and without ventilator-associated pneumonia (VAP)
P aeruginosa virulence determinants during colonisation and VAP
Virulence phenotypes fluctuate during colonisation.26 We therefore measured both elastase and rhamnolipid production in the 320 daily isolates from the 29 patients. According to the daily elastase production, we identified three distinct patterns. Fifteen patients were consistently colonised by low elastase-producing isolates (figure 3A,B). The average daily elastase production value in this group fluctuated between 1% and 52% of the reference strain PAO1. Eight patients were colonised by isolates producing fluctuating intermediate amounts of elastase. This category displayed a clear tendency to decreasing daily average elastase production from 195% (D0) to 24% (D11) (figure 3A,B). Six patients were colonised during their entire observation period by isolates producing high levels of elastase, with average values between 102% and 146% (figure 3A,B). The six patients with VAP (13111, 13128, 16101, 21107, 24101 and 13116) were equally distributed in all three categories, suggesting no association between elastase production of the colonising isolates and VAP. The average per patient elastase production was not different between the VAP and non-VAP groups.
Elastase production of colonising isolates from 29 patients. (A) Elastase production is expressed as a percentage of the reference strain PAO1. Patients were allocated to three different classes according to the temporal evolution of elastase production of their isolates (high, variable and low). The average (AVG) for each group is shown. Patients who developed ventilator-associated pneumonia (VAP) are shown in bold. (B) Temporal evolution of average elastase production of isolates from the three different classes shown in A. Evolution is shown only up to day 11 due to low sample sizes thereafter.
The analysis of the daily rhamnolipid production levels also allowed separation of the 29 patients into three distinct groups (figure 4A,B). Over the study period, 11 patients remained colonised by non-rhamnolipid-producing isolates, with daily average values between 0 and 20% of the reference strain PAO1 (figure 4A,B). Thirteen patients were colonised by isolates producing intermediate amounts of rhamnolipids, with daily average values between 59% and 76% (figure 4A,B). Five patients (13111, 13128, 16101, 21107, 24101) were colonised during the entire study period by isolates producing high rhamnolipid levels, with average daily values between 76% and 100% of the PAO1 reference strain (figure 4A,B). All these five patients developed VAP. The sixth patient with VAP (13116), colonised by a lasR/rhlR double mutant, developed pneumonia in the absence of detectable rhamnolipid production. Thus VAP occurred more frequently in the group of patients colonised during the entire study period by isolates producing high rhamnolipid levels than in the other two groups (p=0.001). Even when patient 13116 was included in the analysis, patients with VAP were colonised over time with isolates producing higher average levels of rhamnolipids than patients without VAP (Mann–Whitney test, p=0.014) (figure 4C).
Rhamnolipid production of colonising isolates from 29 patients. (A) Rhamnolipid production is expressed as a percentage of the reference strain PAO1. Patients were allocated to three different classes according to the temporal evolution of rhamnolipid production of their isolates (high, intermediate and low). The average (AVG) for each group is shown. Patients who developed ventilator-associated pneumonia (VAP) are shown in bold. (B) Temporal evolution of average rhamnolipid production of isolates from the three different classes shown in A. Evolution is shown only up to day 11 due to low sample sizes thereafter. (C) Comparison of average rhamnolipid production per patient during their entire observation period between the VAP (n=6) and non-VAP group (n=23).
‘In patient’ QS induction
To obtain a global picture of QS activity in vivo, we analysed P aeruginosa QS gene expression at the population level by extracting total genomic DNA and total RNA from daily tracheal aspirates (figure 1A). As QS is cell density dependent, we determined the global bacterial lung loads using the genomic DNA. In all patients, the daily bacterial load fluctuated within a range of 106–108 genomic copies/g of aspirate without a clear trend (figure 1B). Except for a transient two- to threefold increase in patients with VAP between the second and fourth day of colonisation, no difference in bacterial load was observed between those with and without VAP (figure 1B). Since all VAP cases (indicated by arrows) occurred on or before day 11, a comparison beyond this point was not possible.
We assessed the expression of two transcriptional regulator genes (lasR and rhlR), and of two autoinducer synthases genes (lasI and rhlI), which together form the QS circuit genes of P aeruginosa, as well as of two major QS target genes lasB (elastase) and rhlA (rhamnosyltransferase).4 We detected expression of lasI and rhlA in 70% and 42%, respectively, of all aspirates (see online data supplement). The expression of lasB and rhlI was measured at lower levels and in fewer samples, excluding these genes from longitudinal analysis. We therefore analysed the mean lasI (figure 5A) and rhlA (figure 5B) expression levels over time within 19 and 16 patients without VAP, respectively, from whom adequate samples for QS gene expression were available using the QS-independent housekeeping gene rpsl to normalise gene expression between samples. Both curves showed a similar stable non-induced profile compatible with basal gene expression. Adequate samples for QS gene expression profiles were available for four patients with VAP (13111, 13116, 13128 and 21107). Patients 13111 and 21107 with VAP showed clear QS induction profiles for both lasI (figure 5A) and rhlA (figure 5B) with an exponential increase of QS gene expression starting 48 h before VAP. Expression levels of patient 13128 with VAP remained comparable with means of patients without VAP. Consistent with the low rhamnolipid production of its colonising isolates (figure 4A), patient 13116 with VAP colonised by a double lasR/rhlR mutant did not show any QS induction.
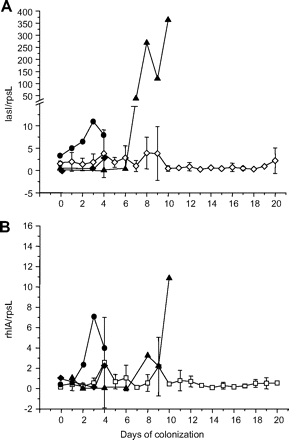
’In patient‘ quorum sensing (QS) induction. (A) Temporal evolution of lasI expression. Three patients with ventilator-associated pneumonia (VAP): 13111 (filled circles), 13128 (filled diamonds) and 21107 (filled triangles), and 19 patients without VAP (open diamonds). (B) Temporal evolution of rhlA expression. Three patients with VAP: 13111 (filled circles), 13128 (filled diamonds) and 21107 (filled triangles), and 16 patients without VAP (squares). Gene expression is standardised towards expression of the housekeeping gene rpsL.
To highlight the previously described cell density dependency and hierarchy of QS gene induction, we measured the expression of the QS circuit (lasR, lasI, rhlR and rhlI) and QS target (lasB and rhlA) genes during in vitro growth of the reference strain PAO1 (figure 6A).27 As expected QS induction occurred only once the bacterial cell density had reached a threshold level (1×109 CFU/ml under our conditions), and the fold induction of the QS target genes lasB and rhlA was higher than that of the QS regulator genes lasR and rhlR. In patient 13111 with VAP we were able to measure the expression of the same QS target and circuit genes over time (figure 6B). In agreement with in vitro data, the rhl QS system genes were more induced than those of the las system in this patient.28 We observed the highest induction in gene expression (100-fold) for the QS target gene rhlA. In contrast to the in vitro situation, the bacterial load in the tracheal aspirates did not increase during the observation period (see inset in figure 6B).
In vitro and ‘in patient’ quorum sensing (QS) induction. (A) In vitro QS circuit induction. Growth curve of the P aeruginosa wild-type strain PAO1 in LB medium (figure inset). Expression (fold change of cDNA copies/ml culture) of QS circuit genes (lasR, rhlR, lasI and rhlI), QS target genes (rhlA and lasB) and the housekeeping gene (rpsL) determined by quantitative reverse tanscription–PCR (qRT–PCR). (B) ‘In patient’ QS circuit induction. Daily QS gene expression of patient 13111 expressed as fold change of cDNA copies/g of aspirate with respect to day 1. Bacterial cell density (inset).
Discussion
This is the first study to report on QS-dependent virulence in P aeruginosa isolates in vitro at the clonal, as well as ‘in patient’ at the bacterial population level, during both colonisation and development of VAP. We found that (1) the majority of patients were initially colonised by QS-deficient P aeruginosa isolates; (2) VAP occurred more frequently in patients colonised by QS-proficient isolates; (3) the production of rhamnolipids, both by the initial colonising isolate and the subsequent colonisers, was associated with development of VAP; and (4) ‘in patient’ QS gene induction was observed only in patients developing VAP.
Our study included 29 colonised patients of which six developed VAP. Only 24% of our patients were initially colonised by fully QS-proficient P aeruginosa isolates. The incidence of VAP was much higher in this group (57%) as compared with the rest of the study population (9%). Moreover all patients initially and persistently colonised during the entire observation period by isolates producing high levels of rhamnolipids developed VAP, supporting the assertion that this particular QS-dependent virulence trait plays a role in the development of this infection. Several mechanisms by which rhamnolipids can promote infection have been proposed. Rhamnolipids are necessary and sufficient for invasion of a reconstituted human airway epithelium; they modulate biofilm architecture and lyse polymorphonuclear neutrophils.29–31 Taken together these data designate rhamnolipids as an essential factor promoting the early stages of mucosal invasion. However, because of the low patient number, our data do not exclude that other QS-dependent factors also play a role during colonisation and infection in intubated patients. Strikingly we did not find a direct association between elevated elastase production and the occurrence of VAP. However, we have shown previously that VAP occurs earlier in patients colonised by elastase-producing isolates, as compared with patients colonised by isolates with reduced or no elastase production.26 Markedly, while rhamnolipid production remained fairly stable over time, average elastase production decreased. This is in agreement with a constant proportion of rhlR mutants (rhl mainly regulating rhamnolipid production) but an increasing proportion of lasR mutants (lasR mainly regulating elastase production) colonising the patients over time.26
While the importance of QS-dependent virulence for pathogenesis has been established in different host models and is further supported by our in vitro data, its relevance needed to be further investigated directly in the human host.9 Our study provides the first ‘in patient’ view of QS dynamics. In patients without VAP, lasI and rhlA were only expressed at a basal, non-induced level during colonisation. In contrast, we observed global QS induction in two colonised patients progressing towards VAP. Although the number of VAP cases is not sufficient to achieve a statistically significant association with ‘in patient’ QS induction, this observation illustrates QS activity in vivo during development of VAP. As ‘in patient’ gene expression data reflect the mean of a global bacterial population, QS induction in spatially restricted areas such as microcolonies, where VAP might start initially, could potentially be missed. This could explain why we were unable to detect QS induction in the other patients with VAP.
In one patient, colonised by QS-proficient isolates, we observed a similar ‘in patient’ QS gene expression profile and hierarchy to that in vitro, illustrating that QS can also orchestrate P aeruginosa gene expression in the lungs of the human host. This ‘in patient’ QS induction occurred without an increase in bacterial cell density and coincided with the development of pneumonia. In contrast, one patient with VAP was colonised by a double lasR/rhlR mutant. Interestingly, isolates of this patient carried the exoU gene, encoding a phospholipase whose expression has been associated with increased mortality from VAP.32 Since QS negatively regulates TTSS expression, these isolates might have achieved higher or earlier expression of TTSS, and development of VAP might have been mediated by cytotoxicity in this particular case.33 Whether colonisation with exoU isolates per se is a risk factor for progression from colonisation to VAP has not been investigated, but our results are not in favour of this hypothesis.
In conclusion, this study shows that the production of rhamnolipids by colonising isolates is associated with the development of VAP in intubated patients. Further studies of the potential benefit of inhibiting this QS-dependent virulence factor in colonised intubated patients are warranted. Macrolide antibiotics, furanone derivatives and N-acylhomoserine (HSL) analogues have been shown in vitro to reduce QS gene expression and the production of QS-dependent virulence factors, and might be good candidates as QS inhibitors.23 34 35 Such studies would provide the scientific basis for bacterial virulence inhibition to prevent P aeruginosa VAP in intubated patients.
Acknowledgments
We express our gratitude to the responsible physicians, especially B Francois, the nurses and medical staff involved in the collection of clinical samples and to F. Brunner-Ferber. We thank R Comte and J L Dumas for outstanding technical assistance.
References
Supplementary materials
Web Only Data thx.2009.133082
Files in this Data Supplement:
Footnotes
Presented in part at the Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC), Washington, DC, 25–28 October 2008 (abstract A 2521).
Funding This work was supported by the Swiss National Science Foundation (grants 4049-063239 to TK and CVD, 320000-108106 and 320000-120011 to CVD), and partially by the Research Fund of the Department of Internal Medicine of the University Hospital and the Faculty of Medicine of Geneva; this Fund receives an unrestricted grant from AstraZeneca Switzerland, Pfizer Switzerland and the Wilsdorf Foundation. The clinical collection was financed by Anbics Corporation.
Competing interests None.
Ethics approval This study was conducted with the approval of the Ethics Committee of the Department of Medicine of the University Hospital Geneva and thereafter from all local ethical committees.
Provenance and peer review Not commissioned; externally peer reviewed.